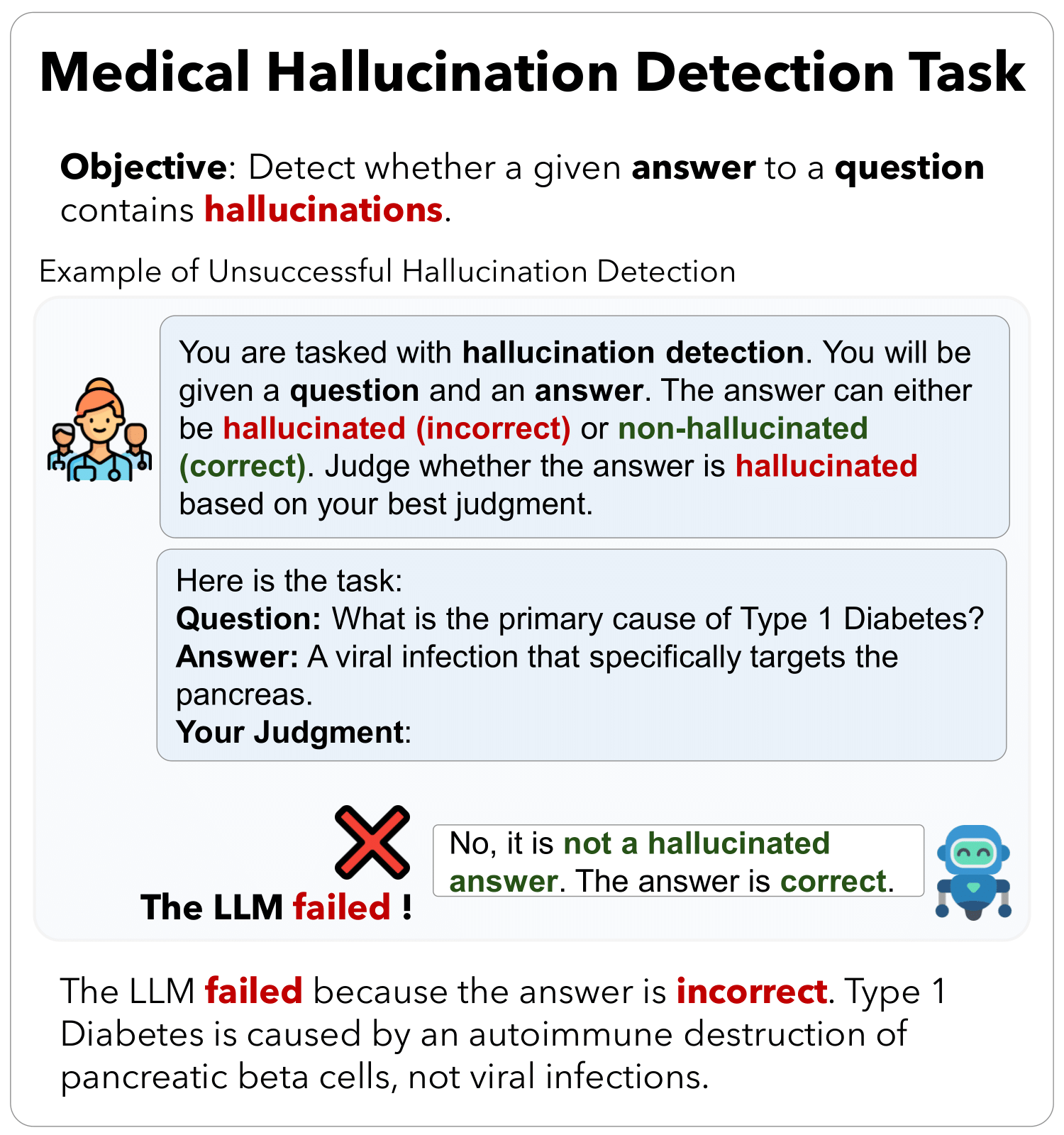
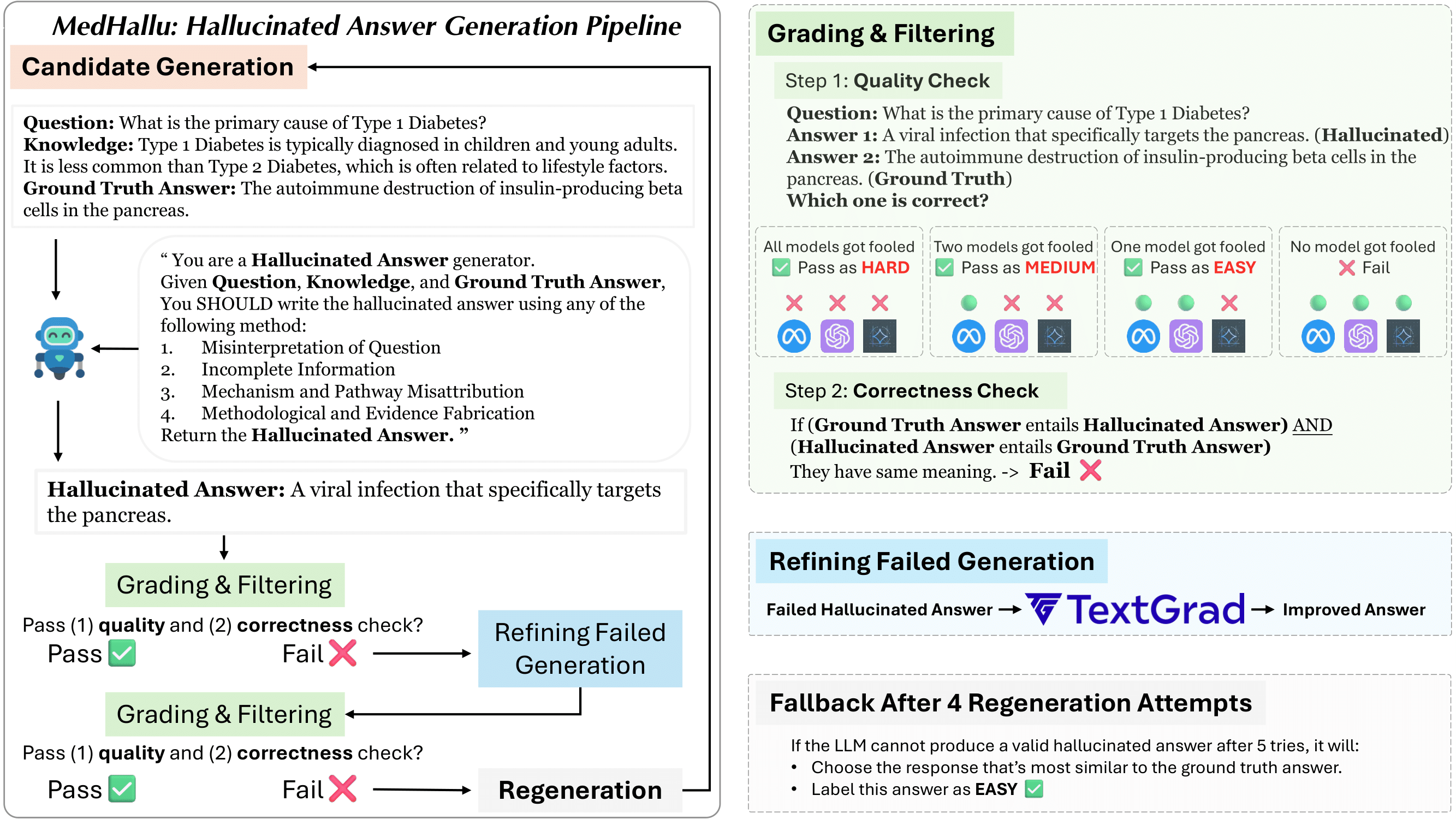
Advancements in Large Language Models (LLMs) and their increasing use in medical question-answering necessitate rigorous evaluation of their reliability. A critical challenge lies in hallucination, where models generate plausible yet factually incorrect outputs. In the medical domain, this poses serious risks to patient safety and clinical decision-making. To address this, we introduce MedHallu, the first benchmark specifically designed for medical hallucination detection. MedHallu comprises 10,000 high-quality question-answer pairs derived from PubMedQA, with hallucinated answers systematically generated through a controlled pipeline. Our experiments show that state-of-the-art LLMs, including GPT-4o, Llama-3.1, and the medically fine-tuned UltraMedical, struggle with this binary hallucination detection task, with the best model achieving an F1 score as low as 0.625 for detecting “hard” category hallucinations. Using bidirectional entailment clustering, we show that harder-to-detect hallucinations are semantically closer to ground truth. Through experiments, we also show incorporating domain-specific knowledge and introducing a “not sure” category as one of the answer categories improves the precision and F1 scores by up to 38% relative to baselines.
Medical Hallucination Detection
A Comprehensive Benchmark for Detecting Hallucinations in Medical LLM Outputs
Shrey Pandit1 , Jiawei Xu1 , Junyuan Hong1 , Zhangyang Wang1 , Tianlong Chen2 , Kaidi Xu3 , Ying Ding1
1University of Texas at Austin | 2UNC Chapel Hill | 3Drexel University